Clinical research organizations
for CROs and their sponsors
Packaged translation and language solutions for scaling clinical trials across languages and geographies
Linguamatics pre-set and proven clinical translation delivery model enables faster recruitment of participants, quicker data analysis, and more timely regulatory submissions, ultimately expediting the development and approval of life-saving treatments and therapies. Available on demand and in any languages.
End-to-end solution for localizing and validating COAs from and to any language and culture, as well as ensuring proper migration on eCOA formats.
Onboard all employees who require translations on a safe and data compliant environment, while reducing rising costs in managing multilingual content. Equip all your operations with the ability to scale across languages and cultures safely, at the push of a button.

Linguamatics designed its technology suite with clinical trials in mind. Our purpose is to offer an off-the-shelf solution to scale clinical trials across any language, geography and regulatory barrier, faster, simpler and more cost efficiently.
Get in touch with a Linguamatics expert to find out how your organization can now provide sponsors with best in class language solutions for their global trials, while reducing costs on their end.
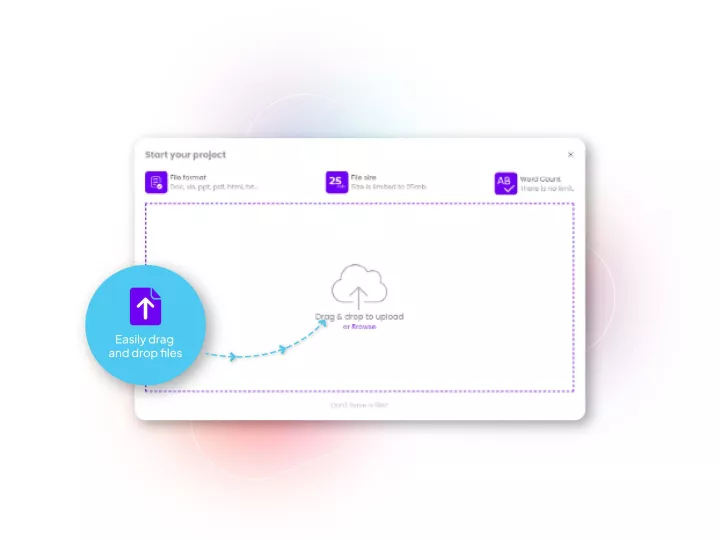
Linguamatics hub is our translation management system designed for clinical trials to maximize speed, quality and integration capabilities while ensuring end-to-end data compliance. It allows all your operations and teams that requires language solutions to obtain them in a few clicks, while maintaining strict compliance and reducing costs.
It integrates in all your system and can be rolled out for your sponsors’ teams or on site teams to access simply and on demand.
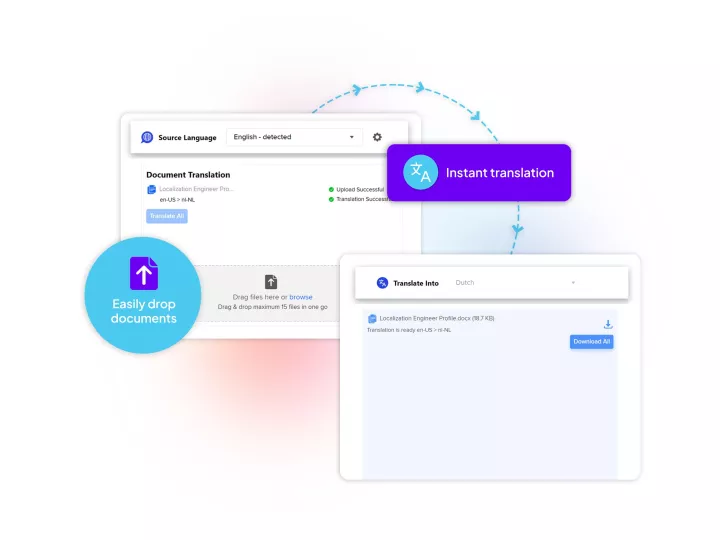
Linguamatics translate is our secured and compliant instant translation system allowing all your employee and system to communicate in any language, securely, for some applications such as:
Linguamatics NLP is our proprietary Natural Language Processing technology developed for life sciences' use cases and enabling unmatched efficiencies in data mining for clinical research and operations.

Choosing an LSP partner that exclusively specializes in servicing the life science industry offers significant advantages:
This focused approach not only brings speed and cost efficiency but also instills confidence in maintaining data compliance and confidentiality throughout the translation process.
